Conventional study of chemical reactions tracks reactant concentrations. This is not a problem if the system of interest contains large number of molecules because all the information needed to describe it is effectively condensed in concentration values. For systems with small number of molecules, concentration values are not descriptive enough. This issue is of particular importance in biochemistry because the number of molecules inside a cell is certainly small.
In these cases, chemical reactions cannot be interpreted as differential equations followed by reactant concentrations. Instead, chemical reactions are interpreted as what they actually are, namely the conversion of a molecule or groups of molecules into different ones. Since occurrence of molecular reactions is largely determined by chance, e.g., a fortuitous encounter of reactants, stochastic modeling is warranted. Further note that it is reasonable to expect the probability of a particular reaction to depend only on the current numbers of molecules for each reactant. Therefore, if we regard numbers of molecules as the state of a stochastic system, chemical reactions can be reasonably modeled as a continuous time Markov chain (CTMC).
This idea is widely used to simulate chemical reactions. The rest of this page covers the following topics
- Lotka-Volterra predator-prey model. A simple example of how chemical reactions are modeled using differential equations.
- Gillespie’s algorithm. Discuss stochastic modeling of Lotka-Volterra’s equations and introduce the workhorse Gillespie’s algorithm.
- Dimerization kinetics. Back and forth conversion between the a molecule and its dimer.
- Enzymatic reactions. Enzymes mediate the production of molecules from reactant components.
- A simple auto-regulatory gene network. An auto-regulatory process to control the amount of protein transcribed from a particular gene.
- Lac operon. The mechanism to balance the digestion of glucose and lactose.
Matlab code for the examples discussed below is in this compressed folder. In the following we make references to files in this folder. Slides containing figures and brief explanations are here. Matlab code and slides were developed by Cameron Finucane during the summer of 2009.
Lotka-Volterra predator-prey model
Lotka and Volterra’s model involves a prey species ![]() and a predator species
and a predator species ![]() . The reactions that define the model describe the birth and death of prey and predator elements. As shorthand notation for the model we use
. The reactions that define the model describe the birth and death of prey and predator elements. As shorthand notation for the model we use
The first reaction ![]() denotes spontaneous reproduction of prey
denotes spontaneous reproduction of prey ![]() . Reaction
. Reaction ![]() denotes consumption of a prey molecule
denotes consumption of a prey molecule ![]() to generate a copy of the predator molecule
to generate a copy of the predator molecule ![]() . Reaction
. Reaction ![]() denotes spontaneous death of predators
denotes spontaneous death of predators ![]() . This is a very simple model but suffices for illustration purposes. Notice that the reactions as summarized in
. This is a very simple model but suffices for illustration purposes. Notice that the reactions as summarized in ![]() –
–![]() are not enough to describe the dynamic behavior of the system for which we need to specify the rate at which the reactions occur. Therefore, we need to specify rates
are not enough to describe the dynamic behavior of the system for which we need to specify the rate at which the reactions occur. Therefore, we need to specify rates ![]() ,
, ![]() , and
, and ![]() that determine how likely a reaction is to occur.
that determine how likely a reaction is to occur.
Classical chemodynamics starts rewriting the above equations to describe the rate of change of predator and prey concentrations. Let ![]() denote the concentration of prey and
denote the concentration of prey and ![]() the concentration of predator at time
the concentration of predator at time ![]() . The rates of change in predator and prey concentration are given by the time derivatives
. The rates of change in predator and prey concentration are given by the time derivatives ![]() and
and ![]() . The concentration of prey
. The concentration of prey ![]() increases when reaction
increases when reaction ![]() occurs and decreases when reaction
occurs and decreases when reaction ![]() occurs. Considering that
occurs. Considering that ![]() happens at a rate
happens at a rate ![]() and
and ![]() at a rate
at a rate ![]() the rate of change
the rate of change ![]() in prey concentration is
in prey concentration is
Likewise, predator numbers increase upon reaction ![]() and decrease with reaction
and decrease with reaction ![]() . Because these reactions happen at rates
. Because these reactions happen at rates ![]() and
and ![]() it follows that
it follows that
There are reasons to expect reaction rates ![]() to be proportional to the concentration of reactants involved in the reaction. That is, we expect
to be proportional to the concentration of reactants involved in the reaction. That is, we expect ![]() to be proportional to the prey’s concentration
to be proportional to the prey’s concentration ![]() ,
, ![]() proportional to the product of both concentrations
proportional to the product of both concentrations ![]() and
and ![]() and
and ![]() to be proportional to the predator’s concentration
to be proportional to the predator’s concentration ![]() . Regardless of what is reasonable to expect there is abundant empirical evidence that this is true. Substituting these expressions into the rate change equations above we obtain the pair of differential equations
. Regardless of what is reasonable to expect there is abundant empirical evidence that this is true. Substituting these expressions into the rate change equations above we obtain the pair of differential equations
The above system of ordinary differential equations (ODE) are the Lotka-Volterra’s ODEs. To simulate a deterministic Lotka-Volterra system use the code in the file ssas_lotka_volterra_deterministic.m.
Gillespie’s algorithm
The differential equations in the previous section are accurate if predator and prey numbers can be accurately inferred from respective concentrations. As expressed before this is not true if the number of reactants is small. In such case randomness might yield distinctive behaviors that can only be captured through a stochastic model. To introduce such stochastic models reconsider the prey-predator model
where Reaction ![]() denotes spontaneous reproduction of prey
denotes spontaneous reproduction of prey ![]() ,
, ![]() consumption of a prey molecule
consumption of a prey molecule ![]() to generate a copy of the predator molecule
to generate a copy of the predator molecule ![]() and
and ![]() spontaneous death of predators
spontaneous death of predators ![]() . To complete the description we interpret
. To complete the description we interpret ![]() and
and ![]() as the number of molecules of each species and introduce hazard functions
as the number of molecules of each species and introduce hazard functions ![]() ,
, ![]() , and
, and ![]() .
.
The definition of the hazard function ![]() is as follows. Given that the number of molecules are
is as follows. Given that the number of molecules are ![]() and
and ![]() at time
at time ![]() , the probability of reaction
, the probability of reaction ![]() occurring in an infinitesimal time interval
occurring in an infinitesimal time interval ![]() is given by
is given by ![]() . As a consequence of this definition, if no other reaction occurs, it follows that the time
. As a consequence of this definition, if no other reaction occurs, it follows that the time ![]() that elapses until the next occurrence of reaction
that elapses until the next occurrence of reaction ![]() is exponentially distributed with parameter
is exponentially distributed with parameter ![]() . This is a basic property of continuous times Markov chains. Notice, though, that other reactions are likely to occur and that as a consequence, time
. This is a basic property of continuous times Markov chains. Notice, though, that other reactions are likely to occur and that as a consequence, time ![]() is not exponentially distributed. However, note that the probability of any reaction occurring in the infinitesimal time interval
is not exponentially distributed. However, note that the probability of any reaction occurring in the infinitesimal time interval ![]() is given by
is given by ![]() . Thus, upon defining the aggregate hazard
. Thus, upon defining the aggregate hazard ![]() we can model the time
we can model the time ![]() until the next reaction as exponentially distributed with parameter
until the next reaction as exponentially distributed with parameter ![]() . Therefore, in a stochastic model of the above predator-prey relations, times
. Therefore, in a stochastic model of the above predator-prey relations, times ![]() between reactions are exponentially distributed with parameter parameters
between reactions are exponentially distributed with parameter parameters ![]() . Once the inter-reaction time
. Once the inter-reaction time ![]() is determined the reaction type
is determined the reaction type ![]() is determined by drawing a decision variable with probabilities proportional to the ratio
is determined by drawing a decision variable with probabilities proportional to the ratio ![]() ,
, ![]() and
and ![]() .
.
To complete the model we need to specify the reaction hazards ![]() . Reaction
. Reaction ![]() happens spontaneously when a prey molecule reproduces. Assuming that prey molecules act independently it is reasonable to assume that the hazard
happens spontaneously when a prey molecule reproduces. Assuming that prey molecules act independently it is reasonable to assume that the hazard ![]() is proportional to the number of prey molecules
is proportional to the number of prey molecules ![]() . Similarly, Reaction
. Similarly, Reaction ![]() happens upon the chance encounter between a prey and a predator molecule. Notice that there are
happens upon the chance encounter between a prey and a predator molecule. Notice that there are ![]() possible pairs of prey and predator molecules. Thus, hazard
possible pairs of prey and predator molecules. Thus, hazard ![]() is reasonably modeled as proportional to the product of number of molecules
is reasonably modeled as proportional to the product of number of molecules ![]() and
and ![]() . Similarly to
. Similarly to ![]() , reaction
, reaction ![]() happens spontaneously, its hazard therefore modeled as
happens spontaneously, its hazard therefore modeled as ![]() proportional to the number of predator molecules.
proportional to the number of predator molecules.
Implementation of this model yields Gillespie’s algorithm for simulation of chemical reactions. The algorithm’s steps are the following
- Initialize
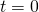 ,
, 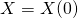 ,
, 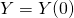
- Calculate all hazards
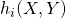
- Find the total hazard,
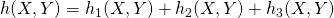
- Find the time to the next reaction:
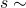 Exp[h(X,Y)]
Exp[h(X,Y)]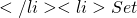 t = t + s
t = t + s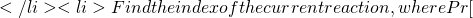 R_i
R_i![Rendered by QuickLaTeX.com ] = h_i(X,Y)/h(X,Y)](https://ese3030.seas.upenn.edu/wp-content/ql-cache/quicklatex.com-180272f374aea211c97468eb90409c42_l3.png)
- Update the state vector to account for this reaction. If reaction
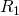 is drawn, update
is drawn, update 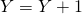 . If reaction 2 occurs update
. If reaction 2 occurs update 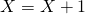 and
and 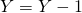 . If
. If 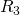 is drawn update
is drawn update 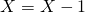 .
. - Repeat from 2
The core steps in the above algorithm are: The random drawing of the next reaction time ![]() (step 4). Determination of the reaction type
(step 4). Determination of the reaction type ![]() (step 6). Update of the reactant numbers
(step 6). Update of the reactant numbers ![]() and
and ![]() (step 7). The remaining steps compute values that are needed to implement these steps. To simulate an stochastic version of the Predator-prey model discussed in the previous subsection use the file ssas_lotka_volterra_stochastic.m. To run this code you need the file ssas_gillespie.m that implements Gillespie’s algorithm for a generically defined chemical equation.
(step 7). The remaining steps compute values that are needed to implement these steps. To simulate an stochastic version of the Predator-prey model discussed in the previous subsection use the file ssas_lotka_volterra_stochastic.m. To run this code you need the file ssas_gillespie.m that implements Gillespie’s algorithm for a generically defined chemical equation.
Dimerization kinetics
A dimer is a molecule composed of two identical (or similar) chemical structures. Each of the pieces of the dimer is called a monomer and can exist in free form. The dimer can separate into its two component monomers and two monomers may combine to form a dimer. The schematic representation of this process is
To complete the model we need to specify either reaction rates ![]() and
and ![]() for a deterministic model or hazards
for a deterministic model or hazards ![]() and
and ![]() for a stochastic framework. Reaction
for a stochastic framework. Reaction ![]() happens spontaneously on dimers. Its hazard is therefore
happens spontaneously on dimers. Its hazard is therefore ![]() proportional to the number of dimer molecules. For reaction
proportional to the number of dimer molecules. For reaction ![]() we have to notice that it happens upon encounter of a pair of monomers. To form a monomer pair we have
we have to notice that it happens upon encounter of a pair of monomers. To form a monomer pair we have ![]() choices for the first member of the pair and
choices for the first member of the pair and ![]() for the second member. The number of ordered pairs is then
for the second member. The number of ordered pairs is then ![]() and the number of unordered pairs is
and the number of unordered pairs is ![]() (there are 2 ordered pairs for each unordered pair). The hazard for reaction
(there are 2 ordered pairs for each unordered pair). The hazard for reaction ![]() is then modeled as
is then modeled as ![]() proportional to the number of unordered monomer pairs. To simulate dimerization kinetics in a deterministic setting use the code in the file ssas_dimerization_deterministic.m. The counterpart code in a stochastic setting is implemented in the file ssas_dimerization_stochastic.m.
proportional to the number of unordered monomer pairs. To simulate dimerization kinetics in a deterministic setting use the code in the file ssas_dimerization_deterministic.m. The counterpart code in a stochastic setting is implemented in the file ssas_dimerization_stochastic.m.
Enzymatic reactions
Enzymes are molecules that mediate chemical reactions. A certain set of reactants can be converted into a certain set of products with some energy expenditure. The enzyme reduces the energy threshold for the reaction to occur. The simplest enzymatic reaction involves the conversion of a substrate ![]() into a product
into a product ![]() . Enzymes
. Enzymes ![]() react with the substrate to generate an intermediate product
react with the substrate to generate an intermediate product ![]() . The intermediate product
. The intermediate product ![]() then liberates the enzyme
then liberates the enzyme ![]() to yield the final product
to yield the final product ![]() . A simple model of this process is given by the following set of reactions
. A simple model of this process is given by the following set of reactions
Reaction ![]() models the combination of a substrate molecule
models the combination of a substrate molecule ![]() and an enzyme
and an enzyme ![]() to yield an intermediate specimen
to yield an intermediate specimen ![]() . The latter may disassociate from the enzyme yielding either the original substrate
. The latter may disassociate from the enzyme yielding either the original substrate ![]() (Reaction
(Reaction ![]() ) or the the final product
) or the the final product ![]() (Reaction
(Reaction ![]() ). Hazards can be determined using the ideas in previous examples. It is ready to realize that hazards can be written as
). Hazards can be determined using the ideas in previous examples. It is ready to realize that hazards can be written as ![]() ,
, ![]() and
and ![]() . Deterministic simulation of enzymatic reactions is in the file ssas_enzyme_deterministic. Stochastic simulation of enzymatic reactions is in ssas_enzyme_stochastic.m.
. Deterministic simulation of enzymatic reactions is in the file ssas_enzyme_deterministic. Stochastic simulation of enzymatic reactions is in ssas_enzyme_stochastic.m.
Auto-regulatory gene network
The three cases studied so far, namely the predator prey model, dimerization kinetics and enzymatic reactions serve as introductory examples, but there are not really interesting differences between the deterministic and stochastic models. Stochastic effects can only be appreciated in full in systems that can be reasonably expected to involve just a handful of specimens.
A good example of this occurs in gene expression. Protein synthesis inside a prokaryotic cell starts with transcription of the corresponding gene into a messenger ribonucleic acid (mRNA). The mRNA then passes through a ribosome to assembly the protein. The number of mRNA molecules synthesized may well be just a few units. The mRNA is an intermediate molecule, the ultimate outcome of this process being the generation of protein. An interesting question is how is the production of protein started and halted at convenient concentrations. The answer to this question varies but in general there are self-regulating feedback loops controlling the production of protein. Roughly speaking, presence of an external stimulus triggers transcription of mRNA and subsequent protein synthesis. As the number of protein molecules increases a byproduct is generated. This byproduct has affinity to bind with the encoding gene thus repressing future transcriptions and halting protein synthesis. A simplified version of an auto-regulatory gene network is the following
| Transcription: | |||
| Assembly: | |||
| Dimerization: | |||
| Repression: | |||
| mRNA degradation: | |||
| Protein degradation: |
Transcription and assembly reactions model the two step protein synthesis. The gene ![]() is transcribed into
is transcribed into ![]() and the
and the ![]() is then used to synthesize protein
is then used to synthesize protein ![]() . Notice that
. Notice that ![]() is not altered upon transcription and that neither is
is not altered upon transcription and that neither is ![]() upon protein synthesis. Auto-regulation is through a dimer
upon protein synthesis. Auto-regulation is through a dimer ![]() of the protein
of the protein ![]() . This dimer generates spontaneously from two protein molecules and may also dissociate spontaneously. This back and forth conversion is denoted by a double arrow in the schematic above. The dimer
. This dimer generates spontaneously from two protein molecules and may also dissociate spontaneously. This back and forth conversion is denoted by a double arrow in the schematic above. The dimer ![]() may attach to the gene
may attach to the gene ![]() . When a dimer is attached to
. When a dimer is attached to ![]() , transcription is not possible and we say that the gene is repressed. We denote this as the generation of the molecule
, transcription is not possible and we say that the gene is repressed. We denote this as the generation of the molecule ![]() . The dimer may spontaneously separate from the gene, making transcription possible once again. This is modeled as the conversion of
. The dimer may spontaneously separate from the gene, making transcription possible once again. This is modeled as the conversion of ![]() into
into ![]() and
and ![]() . In the repression reaction the arrow points in both directions to signify binding and unbinding of the dimer
. In the repression reaction the arrow points in both directions to signify binding and unbinding of the dimer ![]() to the gene
to the gene ![]() . The model is completed by the degradation of mRNA and protein.
. The model is completed by the degradation of mRNA and protein.
This mechanism auto-regulates production of protein ![]() . As the number of protein molecules increases, so does the number of dimers. As the number of dimers increases the likelihood of gene repression increases. We then expect the transcription of mRNA to be halted most of the time. As mRNA molecules degrade the synthesis of protein slows and eventually stops. When this happens, protein degradation reduces the number of dimers. Gene repression becomes less likely, mRNA transcription resumes and consequently so does protein synthesis. The code to simulate this simple auto-regulatory gene network is in the file ssas_autoregulatory_gene_network.m.
. As the number of protein molecules increases, so does the number of dimers. As the number of dimers increases the likelihood of gene repression increases. We then expect the transcription of mRNA to be halted most of the time. As mRNA molecules degrade the synthesis of protein slows and eventually stops. When this happens, protein degradation reduces the number of dimers. Gene repression becomes less likely, mRNA transcription resumes and consequently so does protein synthesis. The code to simulate this simple auto-regulatory gene network is in the file ssas_autoregulatory_gene_network.m.
The lac operon
Auto-regulation in biological systems typically involves group of genes, the proteins they encode for and the byproducts of the reactions of this proteins with other reactants present in the medium. These mechanisms are called auto-regulatory gene networks. One of such networks is the lac operon formed by three adjacent genes that control the metabolism of lactose in some bacteria. Cells can only use glucose to generate energy, but they can reduce lactose to glucose if the latter is unavailable. Such digestion of lactose into glucose occurs in presence of the enzyme ![]() -galactosidase. A simplified model of lactose digestion and glucose consumption is therefore
-galactosidase. A simplified model of lactose digestion and glucose consumption is therefore
| Lactose digestion: | |||
| Glucose consumption: |
where we have use a simplified model of enzymatic reactions (compare with the model in enzymatic reactions. The first reaction models the reduction of lactose ![]() to glucose
to glucose ![]() mediated by the enzyme
mediated by the enzyme ![]() . The second reaction models the consumption of glucose molecules
. The second reaction models the consumption of glucose molecules ![]() .
.
To enable lactose digestion as per the reactions above, cells have to produce the enzyme ![]() -galactosidase, which in itself requires some energy expenditure. Thus, production of
-galactosidase, which in itself requires some energy expenditure. Thus, production of ![]() -galactosidase is only justified when lactose is abundant and glucose scarce. This is controlled by the lac operon.
-galactosidase is only justified when lactose is abundant and glucose scarce. This is controlled by the lac operon.
The constitutive genes on the lac operon are the promoter, the operator and the gene that encodes for ![]() -galactosidase itself. These three genes are adjacent to each other. We note that there are in fact three different kinds of
-galactosidase itself. These three genes are adjacent to each other. We note that there are in fact three different kinds of ![]() -galactosidase, and three corresponding encoding genes, but for the discussion here we can regard them as a single gene. Promoters are integral parts of Deoxyribonucleic acid (DNA) transcription. Transcription of DNA into mRNA necessitates mediation of the enzyme RNA polymerase (RNAP). This enzyme binds to the promoter to initiate transcription of DNA into mRNA. If RNAP does not bind to the promoter, no transcription occurs. The operator is a gene located between the promoter and the lactose encoding sequence. If there is a molecule attached to the operator, it interferes with the action of the enzyme RNAP thereby halting transcription.
-galactosidase, and three corresponding encoding genes, but for the discussion here we can regard them as a single gene. Promoters are integral parts of Deoxyribonucleic acid (DNA) transcription. Transcription of DNA into mRNA necessitates mediation of the enzyme RNA polymerase (RNAP). This enzyme binds to the promoter to initiate transcription of DNA into mRNA. If RNAP does not bind to the promoter, no transcription occurs. The operator is a gene located between the promoter and the lactose encoding sequence. If there is a molecule attached to the operator, it interferes with the action of the enzyme RNAP thereby halting transcription.
A model of this process is then to consider three forms of the the lac operon, the regular operon ![]() , the repressed operon
, the repressed operon ![]() and the activated operon
and the activated operon ![]() . At different rates, transcription of DNA is possible from each of these states, whereby we have the following set of reactions
. At different rates, transcription of DNA is possible from each of these states, whereby we have the following set of reactions
| Repressed transcription: | |||
| Regular transcription: | |||
| Activated transcription: |
The difference in the above reactions is their hazard. Transcription from the repressed operon ![]() is much slower than transcription from the regular operon
is much slower than transcription from the regular operon ![]() resulting in a much smaller hazard. Similartly, for transcription from the activated operon
resulting in a much smaller hazard. Similartly, for transcription from the activated operon ![]() is much larger than the hazard for transcription from the regular operon
is much larger than the hazard for transcription from the regular operon ![]() . Notice that to complete the modeling of
. Notice that to complete the modeling of ![]() -galactosidase production we need to add reactions to model the synthesis of protein from mRNA molecules. These reactions are
-galactosidase production we need to add reactions to model the synthesis of protein from mRNA molecules. These reactions are
| Protein synthesis: | |||
| mRNA degradation: | |||
| Enzyme degradation: |
The first reaction models enzyme synthesis while the other two model degradation of ![]() and
and ![]() .
.
Besides promoter, operator and encoding genes, the control of ![]() -galactosidase production involves a promoter protein and a repressor protein. Upstream of the lac operon is a gene coding for a repressor protein
-galactosidase production involves a promoter protein and a repressor protein. Upstream of the lac operon is a gene coding for a repressor protein ![]() with affinity for the operator gene. This protein is always expressed. When no lactose is present, the repressor binds to the operator thus hindering mRNA transcription and resulting in low
with affinity for the operator gene. This protein is always expressed. When no lactose is present, the repressor binds to the operator thus hindering mRNA transcription and resulting in low ![]() -galactosidase production. When lactose is present, however, the repressor binds preferentially to lactose therefore not interfering with transcription leading to increased production of
-galactosidase production. When lactose is present, however, the repressor binds preferentially to lactose therefore not interfering with transcription leading to increased production of ![]() -galactosidase. The reactions modeling repression of the lac operon are then
-galactosidase. The reactions modeling repression of the lac operon are then
| Operon repression: | |||
| Repressor neutralization: |
The first reaction models the adherence of the repressor protein ![]() to the operon
to the operon ![]() to yield a repressed operon
to yield a repressed operon ![]() . This reaction can be undone as signified by the double arrow above. The second reaction models the binding and unbinding of repressor specimens
. This reaction can be undone as signified by the double arrow above. The second reaction models the binding and unbinding of repressor specimens ![]() to lactose molecules
to lactose molecules ![]() . When there is lactose present we expect the second reaction to occur more frequently than the first. This is not necessarily because the hazard is larger but due to the much larger number of lactose proteins
. When there is lactose present we expect the second reaction to occur more frequently than the first. This is not necessarily because the hazard is larger but due to the much larger number of lactose proteins ![]() with respect to the number of genes
with respect to the number of genes ![]() .
.
The second part of the control involves the catabolite activator protein (CAP) that when bound to the operon facilitates mRNA transcription of ![]() -galactosidase. The amount of CAP present is inversely proportional to the amount of glucose. Hence, when glucose level decreases, the amount of CAP increases and the operon is likely to switch to its activated version. With the operon activated, the rate of mRNA transcription increases and as a consequence so does the production of
-galactosidase. The amount of CAP present is inversely proportional to the amount of glucose. Hence, when glucose level decreases, the amount of CAP increases and the operon is likely to switch to its activated version. With the operon activated, the rate of mRNA transcription increases and as a consequence so does the production of ![]() -galactosidase. The reactions to model this interaction are
-galactosidase. The reactions to model this interaction are
| Operon activation: | |||
| CAP repression: |
The first reaction models the activation of the operon. The second reaction models binding of CAP to glucose ![]() so that the number of free CAP molecules changes in opposite direction to the number of glucose molecules present. We note that CAP does not actually bind to glucose, but for a preliminary model the reactions above suffice.
so that the number of free CAP molecules changes in opposite direction to the number of glucose molecules present. We note that CAP does not actually bind to glucose, but for a preliminary model the reactions above suffice.
When lactose and glucose are present this control mechanism results in a distinctive diauxie pattern with glucose consumed first and lactose processed after glucose is depleted. Simulation of the lac operon is in the file ssas_lac_operon.m.
